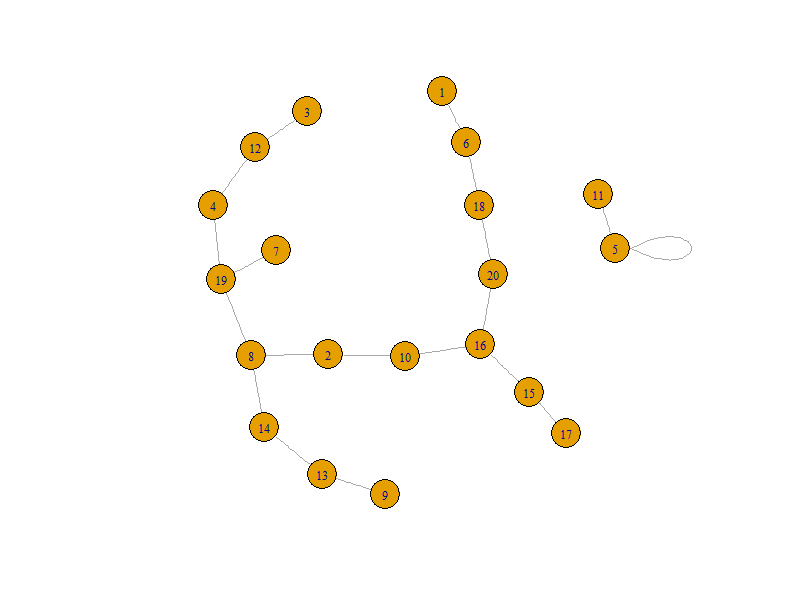
I spent a decent chunk of my morning trying to figure out how to construct a sparse adjacency matrix for use with graph.adjacency(). I’d have thought that this would be rather straight forward, but I tripped over a few subtle issues with the Matrix package. My biggest problem (which in retrospect seems rather trivial) was that elements in my adjacency matrix were occupied by the pipe symbol.
adjacency[1:10,1:10]
10 x 10 sparse Matrix of class 'ngCMatrix'
[1,] . . . . . | . . . .
[2,] . . . . . . . | . .
[3,] . . . . . . . . . .
[4,] . . . . . . . . . .
[5,] . . . . | . . . . .
[6,] . . . . . . . . . .
[7,] . . . . . . . . . .
[8,] . . . . . . . . . .
[9,] . . . . . . . . . .
[10,] . | . . . . . . . .
Of course, the error message I was encountering didn’t point me to this fact. No, that would have been far too simple! The solution is highlighted in the sample code below: you need to specify the symbol used for the occupied sites in the sparse matrix.
library(Matrix)
set.seed(1)
edges = data.frame(i = 1:20, j = sample(1:20, 20, replace = TRUE))
adjacency = sparseMatrix(
i = as.integer(edges$i),
j = as.integer(edges$j),
x = 1,
dims = rep(20, 2),
use.last.ij = TRUE
)
The resulting adjacency matrix then looks like this:
adjacency[1:10,1:10]
10 x 10 sparse Matrix of class 'dgCMatrix'
[1,] . . . . . 1 . . . .
[2,] . . . . . . . 1 . .
[3,] . . . . . . . . . .
[4,] . . . . . . . . . .
[5,] . . . . 1 . . . . .
[6,] . . . . . . . . . .
[7,] . . . . . . . . . .
[8,] . . . . . . . . . .
[9,] . . . . . . . . . .
[10,] . 1 . . . . . . . .
And can be passed into graph.adjacency() without any further issues.
library(igraph)
graph = graph.adjacency(adjacency, mode = 'undirected')