I’ve taken another look at the {hagr} data, which I wrote about previously. This time I’m focusing on the hierarchy of creatures.
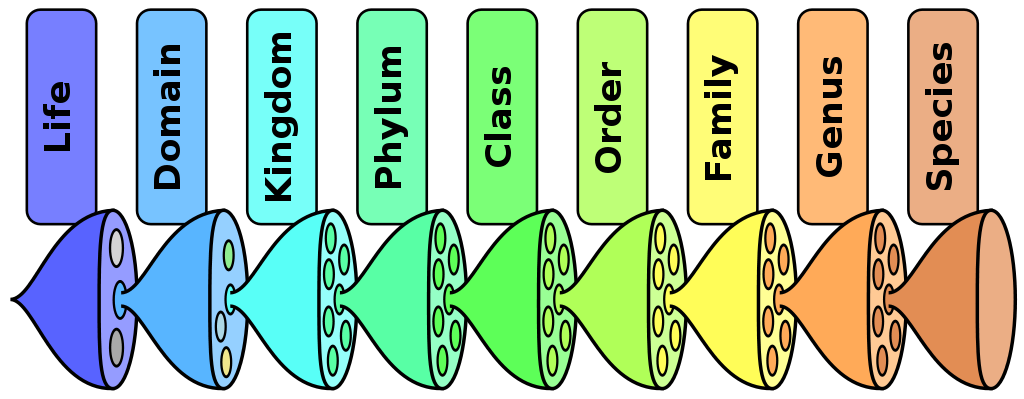
Taxonomic Rank
The Linnaean Taxonomy is a hierarchical classification system for organisms devised by Carl Linnaeus. An organism is assigned to the following levels in the hierarchy (in increasing order or granularity):
- domain
- kingdom
- phylum
- class
- order
- family
- genus and
- species.
The relative level of a group of organisms in this hierarchy determines its taxonomic rank.
💡 The Linnaean Taxonomy was developed way before the idea of evolution arose. As a consequence, despite being a useful framework for classifying organisms, it does not take into account evolutionary relationships.
Let’s take a look at the classification data in the {hagr} package.
library(hagr)
Linnaean Taxonomic Levels
We’ll start at the top level, domain.
age %>% count(domain, sort = TRUE)
# A tibble: 1 × 2
domain n
<chr> <int>
1 Eukarya 4219
There’s only one domain, Eukarya, present. We don’t have any information on Bacteria or Archaea (single-celled organisms).
If we dig down one level then we find that the Eukarya domain consists of three kingdoms: Animalia, Fungi and Plantae. There’s actually a fourth kingdom in Eukarya, Protista, however there’s no data for it in age.
age %>% count(kingdom, sort = TRUE)
# A tibble: 3 × 2
kingdom n
<fct> <int>
1 Animalia 4215
2 Fungi 3
3 Plantae 1
It’s clear that Animalia is the dominant kingdom, so let’s focus on that exclusively.
animalia <- age %>% filter(kingdom == "Animalia")
The next level in the hierarchy is phylum.
animalia %>% count(phylum, sort = TRUE)
# A tibble: 7 × 2
phylum n
<fct> <int>
1 Chordata 4200
2 Arthropoda 8
3 Echinodermata 2
4 Porifera 2
5 Cnidaria 1
6 Mollusca 1
7 Nematoda 1
It appears that Chordata is the dominant phylum in the data, so let’s further narrow our attention.
chordata <- animalia %>% filter(phylum == "Chordata")
Now let’s drill all the way down to genus.
chordata %>% count(class, order, family, genus, sort = TRUE)
# A tibble: 2,035 × 5
class order family genus n
<fct> <fct> <fct> <fct> <int>
1 Teleostei Scorpaeniformes Scorpaenidae Sebastes 49
2 Teleostei Perciformes Percidae Etheostoma 35
3 Aves Passeriformes Parulidae Setophaga 23
4 Teleostei Cypriniformes Cyprinidae Notropis 23
5 Mammalia Chiroptera Vespertilionidae Myotis 21
6 Reptilia Squamata Viperidae Crotalus 19
7 Teleostei Perciformes Lutjanidae Lutjanus 18
8 Aves Psittaciformes Psittacidae Amazona 17
9 Chondrichthyes Carcharhiniformes Carcharhinidae Carcharhinus 17
10 Aves Falconiformes Falconidae Falco 15
# … with 2,025 more rows
Adding in species takes you to the most granular level in the hierarchy.
chordata %>% select(class, order, family, genus, species, common_name)
# A tibble: 4,200 × 6
class order family genus species common_name
<fct> <fct> <fct> <fct> <fct> <chr>
1 Amphibia Anura Bombinatoridae Bombina bombina Firebelly toad
2 Amphibia Anura Bombinatoridae Bombina orientalis Oriental firebelly toad
3 Amphibia Anura Bombinatoridae Bombina variegata Yellow-bellied toad
4 Amphibia Anura Bufonidae Anaxyrus americanus American toad
5 Amphibia Anura Bufonidae Anaxyrus boreas Western toad
6 Amphibia Anura Bufonidae Anaxyrus canorus Yosemite toad
7 Amphibia Anura Bufonidae Anaxyrus cognatus Great plains toad
8 Amphibia Anura Bufonidae Anaxyrus debilis Green toad
9 Amphibia Anura Bufonidae Anaxyrus hemiophrys Canadian toad
10 Amphibia Anura Bufonidae Anaxyrus punctatus Red-spotted toad
# … with 4,190 more rows
💡 The combination of genus and species gives the binomial scientific name for organisms. For example, the Killer Whale is Orcinus orca.
age %>%
filter(str_detect(common_name, "^(Killer|Blue|Sperm) whale$")) %>%
select(class:common_name)
# A tibble: 3 × 6
class order family genus species common_name
<fct> <fct> <fct> <fct> <fct> <chr>
1 Mammalia Cetacea Balaenopteridae Balaenoptera musculus Blue whale
2 Mammalia Cetacea Delphinidae Orcinus orca Killer whale
3 Mammalia Cetacea Physeteridae Physeter macrocephalus Sperm whale
Growing a Tree
We’ll use {ggtree} to construct a phylogenetic tree from domain down to order.
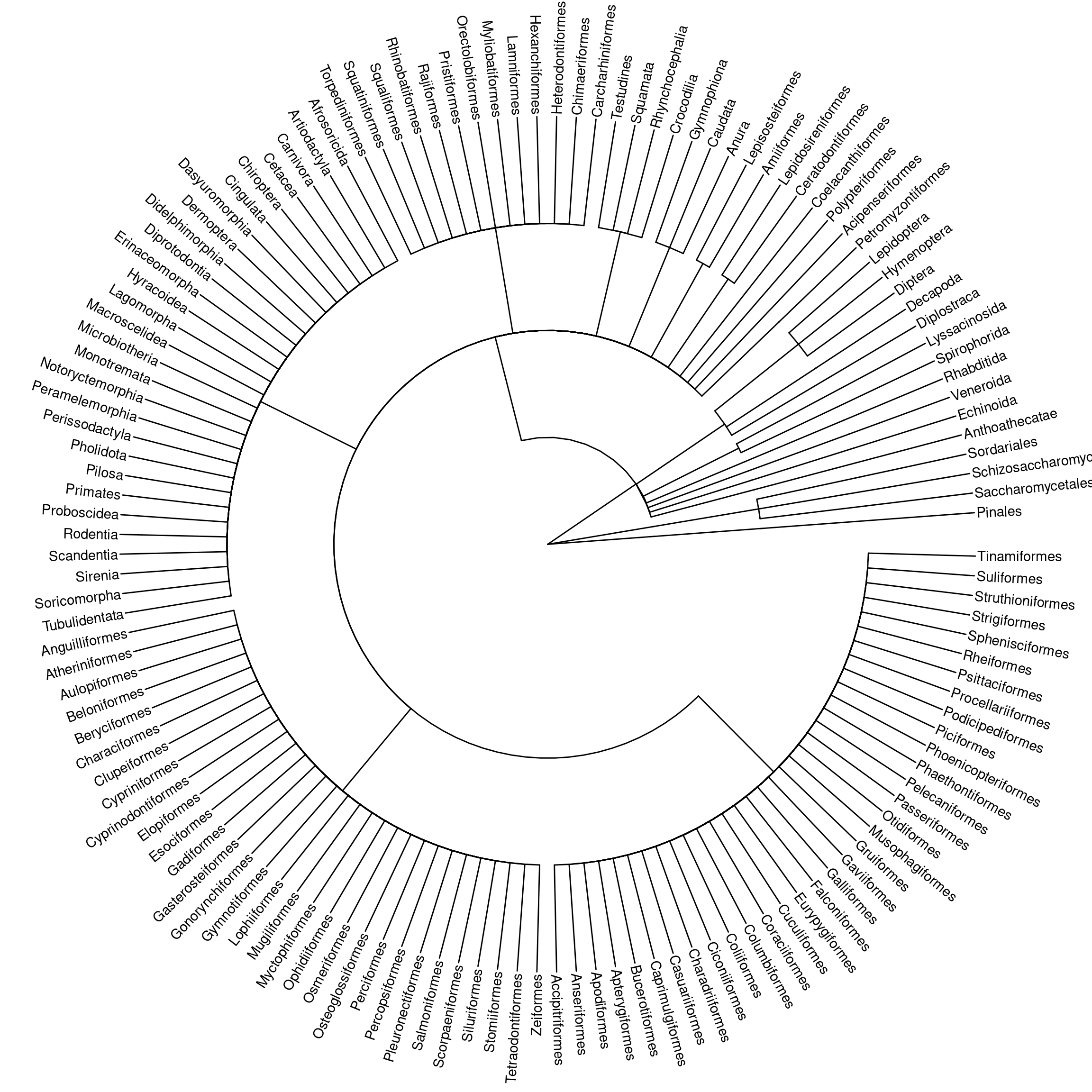
The dominance of the Chordata phylum in the data is readily apparent! It’d be nice to include more levels in this tree, but it gets very big and rather messy.
There’s such a wealth of cool information in this dataset. Really indebted to the Human Ageing Genomic Resources project for putting it together and generously sharing it.